PlaToLoCo is a Platform of Tools for Low Complexity regions in proteins. It can be used to visualise and analyse protein sequences.
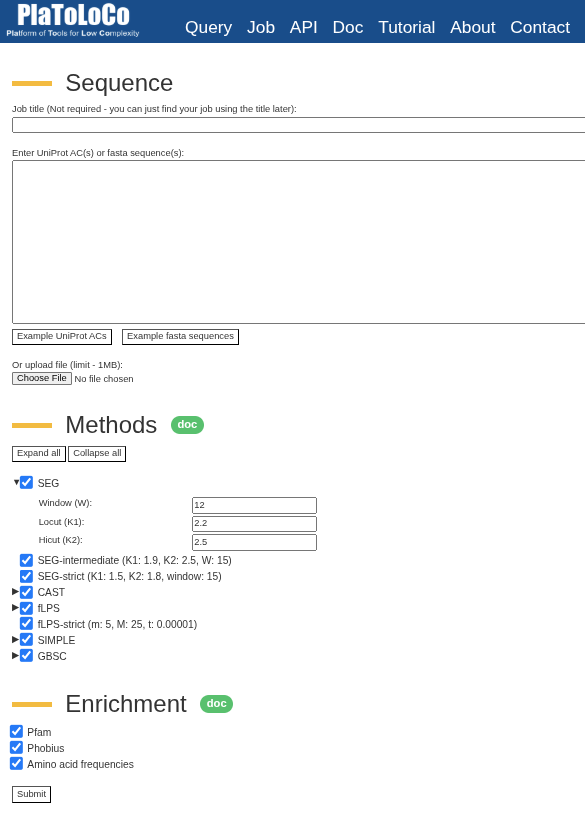
Submit a query
To start, you can go to PlaToLoCo website and fill the query form. Job title you can leave empty if you do not want to find your job in the future. In the second field you can paste your sequences of interest or use “Example UniProt ACs” button to fill the field with example proteins. Alternatively, you can upload a file. In the next section, you can select methods of interest along with their parameters. The “Enrichment” section will include additional data on a feature viewer in the protein detail page. Finally, you can hit the “Submit” button.
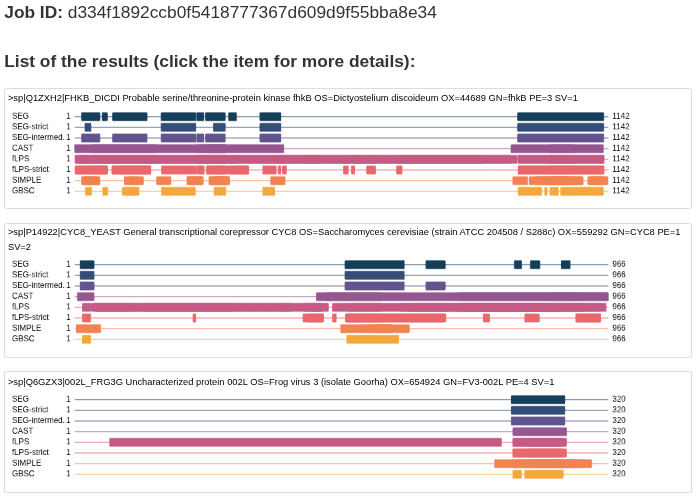
List of proteins
The next page is a list of proteins with places of identified regions. Please note that if a tool identifies two overlapping regions then in this view it is visible as a single long region. For particular regions you need to click an item.
Protein details
This page contain details about a selected protein. It visualize results using feature viewer, amino acid frequency chart, consensus sequence and region details.
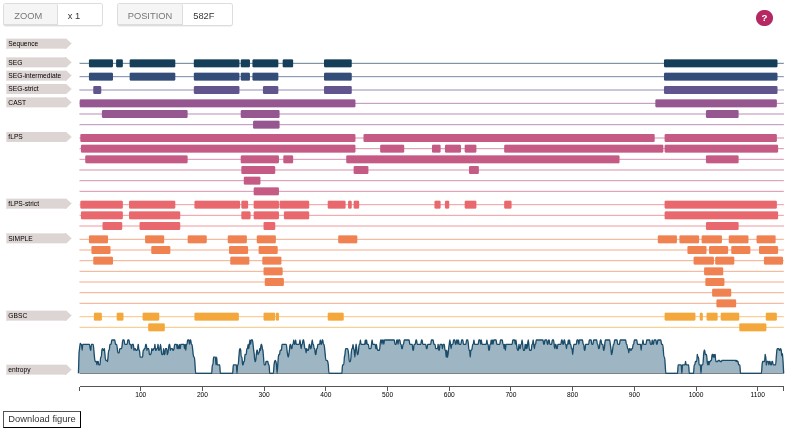
Feature viewer
This view displays selected identification methods with regions identified by each of the method. At the bottom it also shows selected enrichment information and entropy for a sequence.
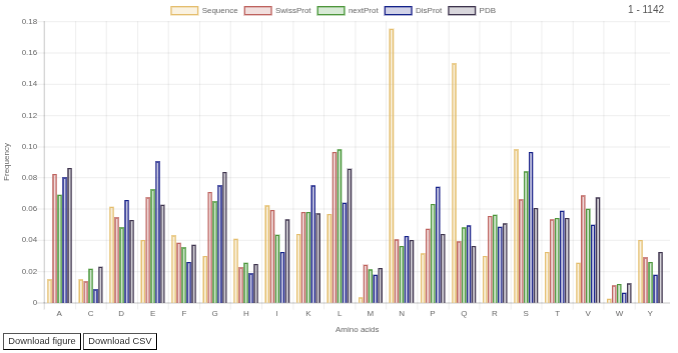
Amino acid frequency
This chart shows amino acid frequency for a sequence and selected databases. If you select a region on the feature viewer then as a sequence it will show amino acid frequency for the selected region.
Sequence consensus
In the consensus view, you can select methods and type to calculate it. Available types are intersection and sum. When intersection is selected then only these amino acids are highlighted which participate in the identified regions in all method. When sum is selected then highlighted are amino acids which were identified by any method.

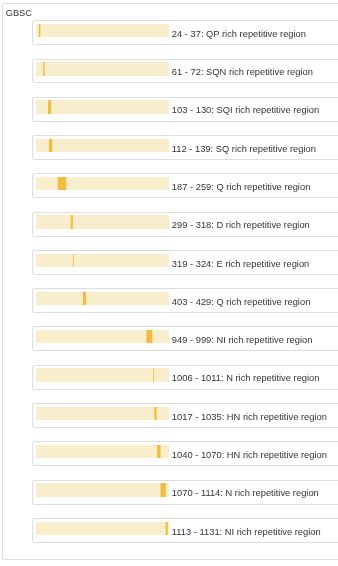
Region details
This view presents list of identified regions along with their location and frequently occurring residues.
How to cite
Jarnot, Patryk, et al. “PlaToLoCo: the first web meta-server for visualization and annotation of low complexity regions in proteins.” Nucleic acids research 48.W1 (2020): W77-W84.